Chronic Kidney Disease Classification
- 15 minsIntroduction
Chronic kidney disease (CKD) is a type of kidney disease in which there is huge loss of kidney function over a long period might be months to years. In this project we used different Machine Learning models and compared between their predictions to get the best classification for the chronic kidney disease.
Steps We took
Preprocessing
Install necessary packages ,Then import libraries.
install.packages(e1071)
install.packages(dplyr)
install.packages(lattice)
install.packages(readxl)
install.packages(rattle)
install.packages(rpart)
install.packages(caret)
install.packages(rpart.plot)
install.packages(caTools)
library(rpart,quietly = TRUE)
library(caret,quietly = TRUE)
library(rpart.plot,quietly = TRUE)
library(rattle)
library(readxl)
library(ggplot2)
library(lattice)
library(caret)
library(dplyr)
library(tidyr)
library(e1071)
library(caTools)
library(class)
Import data
Download data Then import it
dataset <- read.csv('chronic_kidney_disease_full1.csv')Encoding Categorial Data
dataset$sg = factor(dataset$sg, levels= c(1.005,1.01,1.015,1.02,1.025))
dataset$al = factor(dataset$al,levels = c(0,1,2,3,4))
dataset$su = factor(dataset$su,levels = c(0,1,2,3,4,5))
dataset$pc = factor(dataset$pc,levels = c('normal','abnormal'),
labels= c(1,0))
dataset$pcc = factor(dataset$pcc,levels = c('present','notpresent'),
labels= c(1,0))
dataset$ba = factor(dataset$ba,levels = c('present','notpresent'),
labels= c(1,0))
dataset$class = factor(dataset$class, levels = c('ckd','notckd'),
labels = c(1,0))
dataset$rbc = factor(dataset$rbc, levels = c('normal','abnormal'),
labels = c(1,0))
dataset$htn = factor(dataset$htn, levels = c('yes', 'no'),
labels = c(1,0))
dataset$dm = factor(dataset$dm, levels = c('yes', 'no'),
labels = c(1,0))
dataset$cad = factor(dataset$cad, levels = c('yes', 'no'),
labels = c(1,0))
dataset$pe = factor(dataset$pe, levels = c('yes', 'no'),
labels = c(1,0))
dataset$ane = factor(dataset$ane, levels = c('yes', 'no'),
labels = c(1,0))
dataset$appet = factor(dataset$appet, levels = c('good', 'poor'),
labels = c(1,0))
Freature selection
In our data we found that column 21 have alot of missing data so we removed it
dataset <- dataset[,-21]
Data imputation
In chronic_kidney_disease_full1.csv file the missing data is replaced with a ‘?’ so in order to deal with it we had to convert it to ‘NAN’
dataset <- na_if(dataset,'?')
As our data has two types : numeric and catergorical data
We replaced any missing cells with the mean of its feature -if it’s numeric-, Or replaced any missing cells with the mode of its feature -if it’s categorical-.
dataset <- lapply(dataset, function(x)
{
if(is.factor(x)) replace(x, is.na(x), Mode(na.omit(x)))
else if (is.numeric(x)) replace(x, is.na(x) , mean(x, na.rm = TRUE))
else x
})
We must convert the dataset into dataframe to be able to deal with it.
dataset = data.frame(dataset)
Feature normalization
To avoid the increasing in error when the range of the numeric data in each column is different in the width,we attempted feature scalling the data.
dataset [, 1:11] = scale(dataset[,1:11])
Methodology
To increase our accuarcy we used cross validation concepts by creating data folds.
folds = createFolds(dataset$class , k = 5)Used Algorithms
Different algorithms were used to get the most accurate one
1.KNN
2.Tree Decision
3.Logistic Regression
4.Naive Bayes
1. KNN
KNN is the simplest algorithm with a very high accuracy for binary classification, It classifies a new data point based on the similarity.
To apply KNN model we must select K of neighbors after calculating distance between all points and the new data point , It will take the closest k points compared to the new point then it classifies the new point based on the majority. for example: if the closest 5 points 3 of them from A category and 2 from B category ,So this point will be classified as A category.
code
This function will build the model and calculate accuracy, sensitivty and specifity for each fold, Then we put the output in a vector called statistics
cv_KNN = lapply(folds, function(x){
training_fold = dataset[-x,]
test_fold = dataset [x,]
k_nn <- knn(train = training_fold,test = test_fold, cl = dataset[-x,24], k=9)
CM_V = table(dataset[x,24], k_nn)
accuracy_V = (CM_V[1,1] + CM_V[2,2]) / (CM_V[1,1] + CM_V[2,2] + CM_V[1,2] + CM_V[2,1])
sens_v = (CM_V[1,1])/(CM_V[1,1]+CM_V[2,1])
spec_v = (CM_V[2,2])/(CM_V[2,2]+CM_V[1,2])
statisitcs = c(accuracy_V,sens_v,spec_v)
return(statisitcs)
})lapply function will return the output as a list so to deal with it , convert cv_KNN into data frame
cv_KNN <- data.frame(cv_KNN)Then to show the results , Use paste function .
paste("[K-NN] accuracy:",mean(as.numeric(cv_KNN[1,]))
,",sens:" ,mean(as.numeric(cv_KNN[2,]))
,",spec:",mean(as.numeric(cv_KNN[3,])))The results
2. Deision Tree
It is an algorithm used to classify data. This algorithm uses the principle of divide and conquer to divide the problem into parts and solve all of them separately, Then the solution is grouped . The decision tree is created based on the choice of the best attribute, The training set can be divide so that the depth of the tree decreases at the same time as the data is correctly categorized. For more illustration you can check this link
code
cv_dtree = lapply(folds, function(x){
training_fold = dataset[-x,]
test_fold = dataset [x,]
tree <- rpart(class~.,
data= training_fold,
method = "class")
pred <- predict(object=tree,test_fold,type="class")
CM_V = table(pred, test_fold$class)
accuracy_V = (CM_V[1,1] + CM_V[2,2]) / (CM_V[1,1] + CM_V[2,2] + CM_V[1,2] + CM_V[2,1])
sens_v = (CM_V[1,1])/(CM_V[1,1]+CM_V[2,1])
spec_v = (CM_V[2,2])/(CM_V[2,2]+CM_V[1,2])
statisitcs = c(accuracy_V,sens_v,spec_v)
return(statisitcs)
})
cv_dtree <- data.frame(cv_dtree)
paste("[Desicion tree]accuracy:",mean(as.numeric(cv_dtree[1,]))
,",sens:" ,mean(as.numeric(cv_dtree[2,]))
,",spec:",mean(as.numeric(cv_dtree[3,])))
The results
3. Logistic regression
To understand logistic regression you might want to check Linear regression. As you have seen in linear regression , we used a hypothesis relation to predict the output but this time we need that our predictions be the binary outcome of either 0 or 1. So, we use the same hypothesis but with a little modification by using the sigmoid function.
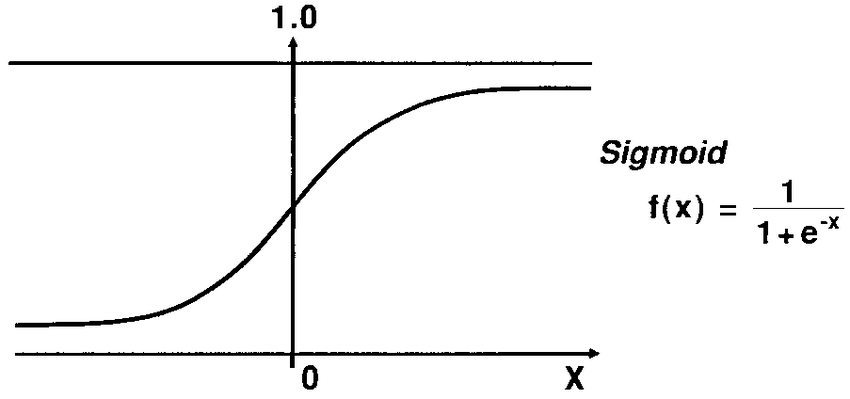
As seen in the figure above , The sigmoid function changed the output into a range between zero and one. To predict whether the output is one or zero ,we need to set a boundary value which is set by default equals to 0.5 , when the output >= 0.5 then it is rounded up to 1 and if it is < 0.5 then it is rounded down to 0. For further details please check : Logistic Regression
Code
cv_LR = lapply(folds, function(x){
training_fold = dataset[-x,]
test_fold = dataset [x,]
classifier = glm (formula =class ~.,
family = binomial,
data = training_fold)
prob_pred = predict.glm(classifier , type = 'response', newdata = test_fold[-24])
y_pred = ifelse(prob_pred > 0.5 , 1 ,0 )
CM_V = table(dataset[x,24], y_pred)
accuracy_V = (CM_V[1,1] + CM_V[2,2]) / (CM_V[1,1] + CM_V[2,2] + CM_V[1,2] + CM_V[2,1])
sens_v = (CM_V[1,1])/(CM_V[1,1]+CM_V[2,1])
spec_v = (CM_V[2,2])/(CM_V[2,2]+CM_V[1,2])
statisitcs = c(accuracy_V,sens_v,spec_v)
return(statisitcs)
})
cv_LR = data.frame(cv_LR)
paste("[LR] accuracy:",mean(as.numeric(cv_LR[1,]))
,",sens:" ,mean(as.numeric(cv_LR[2,]))
,",spec:",mean(as.numeric(cv_LR[3,])))
The results
4. Naive Bayes
This method is based on Bayes Theorem in statistics . It considers two main assumptions ,The first one is that every feature is independent of the other features , The second one is that every feature has the same contribution (Weight) as the others . It calculates the probability of each feature individually given the output and the probability of the output and hence be able to predict the output given the features values . As mentioned in the Logistic regression method we are going to use the same boundary value . For Further details for the algorithm Check Naive Algorithm
Code
cv_naive = lapply(folds, function(x){
training_fold = dataset[-x,]
test_fold = dataset [x,]
Naive_Bayes_Model=naiveBayes(class ~., data= training_fold)
NB_Predictions=predict(Naive_Bayes_Model,test_fold)
CM_V = table(NB_Predictions,test_fold$class)
accuracy_V = (CM_V[1,1] + CM_V[2,2]) / (CM_V[1,1] + CM_V[2,2] + CM_V[1,2] + CM_V[2,1])
sens_v = (CM_V[1,1])/(CM_V[1,1]+CM_V[2,1])
spec_v = (CM_V[2,2])/(CM_V[2,2]+CM_V[1,2])
statisitcs = c(accuracy_V,sens_v,spec_v)
return(statisitcs)
})
cv_naive = data.frame(cv_naive)
paste("[Naive Bayes] accuracy:",mean(as.numeric(cv_naive[1,]))
,"sens:" ,mean(as.numeric(cv_naive[2,]))
,"spec:",mean(as.numeric(cv_naive[3,])))
The results
Conclusion
From the shown results , Although each model acheived a good accuracy , you still need to check for the most suitable model that goes along best with your data.